T Cell Receptors Targeting KRAS Mutants for Cancer Immunotherapy/Adoptive Cell Therapy
Mutations in the Kirsten rat sarcoma viral oncogene homolog (KRAS) gene are among the most common oncogenic drivers in human cancers, affecting nearly a third of all solid tumors. Point mutations in the KRAS gene most frequently affect amino acid position 12, resulting in the substitution of the native glycine (G) residue for other amino acids (e.g., aspartic acid (D), valine (V), cysteine (C) or arginine (R)). The mutations in KRAS occur early in the process of carcinogenesis, and only tumor cells express driver mutations, making them an attractive cancer-specific therapeutic target. However, despite decades of research into the signaling of mutated KRAS and druggability of these mutations with selective inhibitors, no effective therapy has been developed for these common mutated KRAS-driven cancers.
T cell receptors (TCRs) are proteins expressed on the cell surface of T lymphocytes that can recognize peptide antigens from infected and malignant cells in the context of human leukocytes antigen (HLA) molecules with exquisite specificity. Subsequent T cell activation leads to an immune response which aims to eliminate the abnormal cells. T lymphocytes that naturally lack specificity for a tumor antigen can be equipped to express a tumor antigen-specific TCR using genetic engineering. Adoptive transfer of these tumor antigen-specific TCR-engineered T cells into patients with cancer has demonstrated to be a promising cancer treatment strategy.
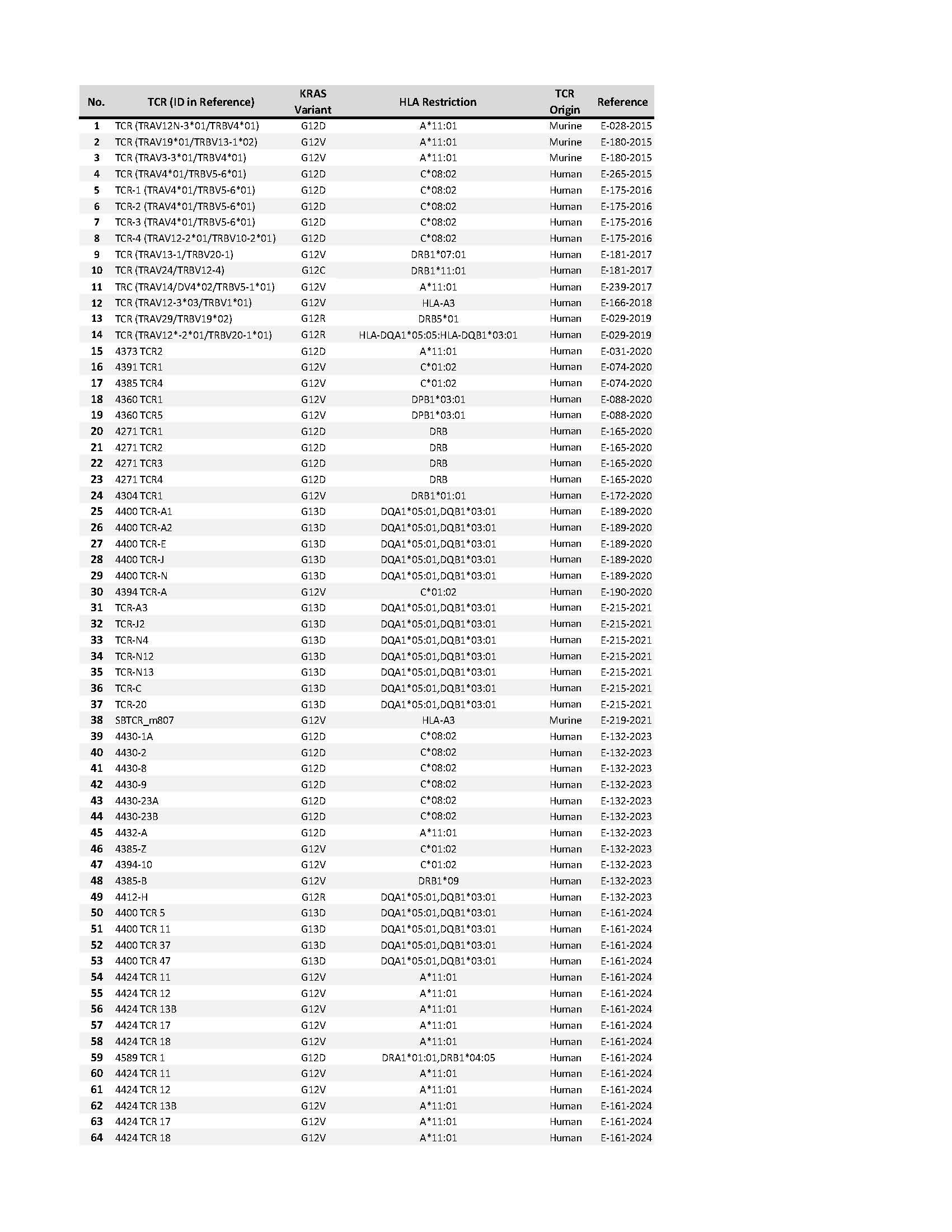
Scientists at the NIH identified a collection of TCRs that specifically recognize mutated KRAS variants (Table 1). These variants cover the most common KRAS driver mutations expressed by a variety of epithelial cancers, including pancreatic, colorectal and lung cancer. The mutated KRAS variants are recognized by the TCRs in the context of specific Class I/Class II HLA alleles. These TCRs are expected to eliminate human cancer cells that express both the appropriate mutated KRAS variant and HLA molecule upon adoptive transfer into patients with cancer. Furthermore, these TCRs can be used for a variety of other experimental therapeutic, diagnostic and research applications.
Table 1: Collection of mutated KRAS TCRs
Table 2: Priority Patent Filings by Invention Reference1.jpg)
Competitive Advantages:
- Highly expressed target antigens: mutated KRAS variants are frequently expressed by the most common epithelial cancers, including pancreatic, colorectal and lung cancer
- Cancer-specific driver mutations: mutated KRAS variants are solely expressed by cancer cells, and not by healthy tissues
- Variety of HLA-restriction elements: extends the applicability of TCRs as they recognize mutated KRAS variants in the context of multiple HLA molecules
Commercial Applications:
- A component of a combination immunotherapy aimed at targeting mutated KRAS-driven cancers
- An in vitro diagnostic tool to screen for cells expressing mutated KRAS in antigen detection assays
- A research tool to investigate signaling in mutated KRAS antigen expressing cells
- Use of portions of the TCRs in chimeric proteins for research and therapeutic purposes in mutated KRAS-driven cancers
Related Inventions
-
E-028-2015
TAB-4010
Novel Cancer Immunotherapy: A T Cell Receptor That Specifically Recognizes Common KRAS Mutations - E-029-2019
- E-031-2020
- E-074-2020
- E-088-2020
- E-105-2012
- E-165-2020
- E-166-2018
- E-172-2020
-
E-176-2014
TAB-4126
T-cell Receptor Targeting Human Papillomavirus-16 E7 Oncoprotein - E-180-2015
-
E-181-2016
TAB-3105
Zika Virus Vaccines - E-189-2020
- E-190-2020
- E-239-2017
- E-265-2015
-
E-495-2013
TAB-3933
T-cell Receptor Targeting Human Papillomavirus-16 E6 Oncoprotein
Patents
- US
Provisional (PRV) 62/369,883
Filed on 2016-08-02
Status: Abandoned - Patent Cooperation Treaty
(PCT) PCT/US2017/044615
Filed on 2017-07-31
Status: Expired - Australia
National Stage 2017306038
Filed on 2017-07-31
Status: Issued - Canada
National Stage 3032870
Filed on 2017-07-31
Status: Pending - China
National Stage 201780059356.4
Filed on 2017-07-31
Status: Issued - European Patent
National Stage 17749580.1
Filed on 2019-01-30
Status: Issued - Japan
National Stage 2019-505220
Filed on 2017-07-31
Status: Issued - US Patent 10,611,816
Filed on 2019-01-30
Status: Issued - Israel
National Stage 264425
Filed on 2019-01-23
Status: Issued - South Korea
National Stage 10-2019-7005837
Filed on 2019-02-27
Status: Issued - Singapore
National Stage 11201900654Q
Filed on 2019-01-24
Status: Issued - Hong Kong
European patent (EP) 19133082.8
Filed on 2019-12-03
Status: Issued - Hong Kong
China Patent (CN) 19132196.7
Filed on 2019-11-14
Status: Issued - Singapore
Divisional (DIV) 10201913959W
Filed on 2019-12-31
Status: Pending - US Patent 11,208,456
Filed on 2020-04-02
Status: Issued - US Patent 11,897,933
Filed on 2021-06-11
Status: Issued - US Patent 11,840,561
Filed on 2021-12-03
Status: Issued - Japan
Divisional (DIV) 2021-199878
Filed on 2021-12-09
Status: Issued - European Patent
Divisional (DIV) 22182473.3
Filed on 2022-07-01
Status: Pending - Albania
European patent (EP) 17749580.1
Filed on 2017-07-31
Status: Issued - Austria
European patent (EP) 17749580.1
Filed on 2017-07-31
Status: Issued - Belgium
European patent (EP) 17749580.1
Filed on 2017-07-31
Status: Issued - Bulgaria
European patent (EP) 17749580.1
Filed on 2017-07-31
Status: Issued - Croatia
European patent (EP) 17749580.1
Filed on 2017-07-31
Status: Issued - Cyprus
European patent (EP) 17749580.1
Filed on 2017-07-31
Status: Issued - Czech Republic
European patent (EP) 17749580.1
Filed on 2017-07-31
Status: Issued - Denmark
European patent (EP) 17749580.1
Filed on 2017-07-31
Status: Issued - Estonia
European patent (EP) 17749580.1
Filed on 2017-07-31
Status: Issued - Finland
European patent (EP) 17749580.1
Filed on 2017-07-31
Status: Issued - France
European patent (EP) 17749580.1
Filed on 2017-07-31
Status: Issued - Germany
European patent (EP) 17749580.1
Filed on 2017-07-31
Status: Issued - Greece
European patent (EP) 17749580.1
Filed on 2017-07-31
Status: Issued - Hungary
European patent (EP) 17749580.1
Filed on 2017-07-31
Status: Issued - Iceland
European patent (EP) 17749580.1
Filed on 2017-07-31
Status: Issued - Ireland
European patent (EP) 17749580.1
Filed on 2017-07-31
Status: Issued - Italy
European patent (EP) 17749580.1
Filed on 2017-07-31
Status: Issued - Latvia
European patent (EP) 17749580.1
Filed on 2017-07-31
Status: Issued - Lithuania
European patent (EP) 17749580.1
Filed on 2017-07-31
Status: Issued - Luxembourg
European patent (EP) 17749580.1
Filed on 2017-07-31
Status: Issued - North Macedonia
European patent (EP) 17749580.1
Filed on 2017-07-31
Status: Issued - Malta
European patent (EP) 17749580.1
Filed on 2017-07-31
Status: Issued - Monaco
European patent (EP) 17749580.1
Filed on 2017-07-31
Status: Issued - The Netherlands
European patent (EP) 17749580.1
Filed on 2017-07-31
Status: Issued - Norway
European patent (EP) 17749580.1
Filed on 2017-07-31
Status: Issued - Poland
European patent (EP) 17749580.1
Filed on 2017-07-31
Status: Issued - Portugal
European patent (EP) 17749580.1
Filed on 2017-07-31
Status: Issued - Romania
European patent (EP) 17749580.1
Filed on 2017-07-31
Status: Issued - Serbia
European patent (EP) 17749580.1
Filed on 2017-07-31
Status: Issued - San Marino
European patent (EP) 17749580.1
Filed on 2017-07-31
Status: Issued - Slovakia
European patent (EP) 17749580.1
Filed on 2017-07-31
Status: Issued - Slovenia
European patent (EP) 17749580.1
Filed on 2017-07-31
Status: Issued - Spain
European patent (EP) 17749580.1
Filed on 2017-07-31
Status: Issued - Sweden
European patent (EP) 17749580.1
Filed on 2017-07-31
Status: Issued - Switzerland
European patent (EP) 17749580.1
Filed on 2017-07-31
Status: Issued - United Kingdom
European patent (EP) 17749580.1
Filed on 2017-07-31
Status: Issued - Turkey
European patent (EP) 17749580.1
Filed on 2017-07-31
Status: Issued - Israel
Divisional (DIV) 301894
Filed on 2023-04-03
Status: Issued - South Korea
Divisional (DIV) None
Filed on None
Status: Administratively Closed
Publications
Collaborations
- Licensing
- Collaboration